Cookbook
In this chapter you can find multiple recipes for different ESM-Tools functionalities, such running a model, adding forcing files, editing defaults in namelists, etc.
If you’d like to contribute with your own recipe, or ask for a recipe, please open a documentation issue on our GitHub repository.
Note
Throughout the cookbook, we will sometimes refer to a nested part of a
configuration via dot notation, e.g. a.b.c. Here, we mean the following
in a YAML config file:
a:
b:
c: "foo"
This would indicate that the value of a.b.c is "foo". In Python, you
would access this value as a["b"]["c"].
Change/Add Flags to the sbatch Call
Feature available since version: 4.2
If you are using SLURM batch system together with ESM-Tools (so far the default
system), you can modify the sbatch call flags by modifying the following variables
from your runscript, inside the computer section:
Key |
Description |
|---|---|
mail_type, mail_user |
Define these two variables to get updates about your slurm-job through email. |
single_proc_submit_flag |
By default defined as |
additional_flags |
To add any additional flag that is not predefined in ESM-Tools |
Example
Assume you want to run a simulation using the Quality of Service flag (--qos) of
SLURM with value 24h. Then, you’ll need to define the additional_flags inside
the computer section of your runscript. This can be done by adding the following to
your runscript:
computer:
additional_flags: "--qos=24h"
Adding more than one flag
Alternatively, you can include a list of additional flags:
computer:
additional_flags:
- "--qos=24h"
- "--comment='My Slurm Comment'"
See the documentation for the batch scheduler on your HPC system to see the allowed options.
Applying a temporary disturbance to ECHAM to overcome numeric instability (lookup table overflows of various kinds)
Feature available since version: esm_runscripts v4.2.1
From time to time, the ECHAM family of models runs into an error resulting
from too high wind speeds. This may look like this in your log files:
30: ================================================================================
30:
30: FATAL ERROR in cuadjtq (1): lookup table overflow
30: FINISH called from PE: 30
To overcome this problem, you can apply a small change to the factor “by which
stratospheric horizontal diffussion is increased from one level to the next
level above.” (mo_hdiff.f90), that is the namelist parameter enstdif,
in the dynctl section of the ECHAM namelist. As this is a common problem,
there is a way to have the run do this for specific years of your simulation. Whenever
a model year crashes due to numeric instability, you have to apply the method outlined
below.
Generate a file to list years you want disturbed.
In your experiment script folder (not the one specific for each run), you can create a file called
disturb_years.dat. An abbreviated file tree would look like:
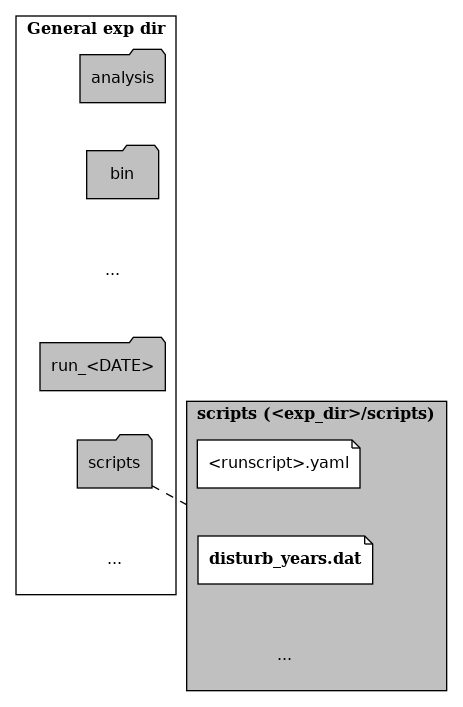
disturb_years.dat location
Add years you want disturbed.
The file should contain a list of years the disturbance should be applied to, seperated by new lines. In practice, you will add a new line with the value of the model year during which the model crashes whenever such a crash occurs.
Example
In this example, we disturb the years 2005, 2007, and 2008 of an experiment
called EXAMPLE running on ollie:
$ cat /work/ollie/pgierz/test_esmtools/EXAMPLE/scripts/disturb_years.dat
2005
2007
2008
You can also set the disturbance strength in your configuration under
echam.disturbance. The default is 1.000001. Here, we apply a 200%
disturbance whenever a “disturb_year” is encountered.
echam:
disturbance: 2.0
See also
ECHAM6 User Handbook, Table 2.4, dynctl
Changing Namelist Entries from the Runscript
Feature available since version: 4.2
You can modify namelists directly from your user yaml runscript configuration.
Identify which namelist you want to modify and ensure that it is in the correct section. For example, you can only modify
ECHAMspecific namelists from anECHAMblock.Find the subsection (“chapter”) of the namelist you want to edit.
Find the setting (“key”) you want to edit
Add a
namelist_changesblock to your configuration, specify next the namelist filename you want to modify, then the chapter, then the key, and finally the desired value.
In dot notation, this will look like:
<model_name>.namelist_changes.<namelist_name>.<chapter_name>.<key_name> = <value>
Example
Here are examples for just the relevant YAML change, and for a full runscript using this feature.
In this example, we modify the co2vmr of the radctl section of
namelist.echam.
echam:
namelist_changes:
namelist.echam:
radctl:
co2vmr: 1200e-6
In this example, we set up AWI-ESM 2.1 for a 4xCO2 simulation. You can see how multiple namelist changes are applied in one block.
general:
setup_name: "awiesm"
compute_time: "02:30:00"
initial_date: "2000-01-01"
final_date: "2002-12-31"
base_dir: "/work/ab0246/a270077/For_Christian/experiments/"
nmonth: 0
nyear: 1
account: "ab0246"
echam:
restart_unit: "years"
nprocar: 0
nprocbr: 0
namelist_changes:
namelist.echam:
radctl:
co2vmr: 1137.e-6
parctl:
nprocar: 0
nprocbr: 0
runctl:
default_output: True
awiesm:
version: "2.1"
postprocessing: true
scenario: "PALEO"
model_dir: "/work/ab0246/a270077/For_Christian/model_codes/awiesm-2.1/"
fesom:
version: "2.0"
res: "CORE2"
pool_dir: "/pool/data/AWICM/FESOM2"
mesh_dir: "/work/ba1066/a270061/mesh_CORE2_finaltopo_mean/"
restart_rate: 1
restart_unit: "y"
restart_first: 1
lresume: 0
namelist_changes:
namelist.config:
paths:
ClimateDataPath: "/work/ba0989/a270077/AWIESM_2_1_LR_concurrent_rad/nonstandard_input_files/fesom/hydrography/"
jsbach:
input_sources:
jsbach_1850: "/work/ba1066/a270061/mesh_CORE2_finaltopo_mean/tarfilesT63/input/jsbach/jsbach_T63CORE2_11tiles_5layers_1850.nc"
Practical Usage
It is generally a good idea to run your simulation once in check mode before actually submitting and examining the resulting namelists:
$ esm_runscripts <your_config.yaml> -e <expid> -c
The namelists are printed in their final form as part of the log during the job
submission and can be seen on disk in the work folder of your first
run_XZY folder.
Note that you can have several chapters for one namelist or several namelists
included in one namelist_changes block, but you can only have one
namelist_changes block per model or component (see
Changing Namelists).
Unusual Namelists
Some times, you have strange namelists of the form:
sn_tracer(1) = 'DET' , 'Detritus ' , 'mmole-N/m3' , .false.
sn_tracer(2) = 'ZOO' , 'Zooplankton concentration ' , 'mmole-N/m3' , .false.
sn_tracer(3) = 'PHY' , 'Phytoplankton concentration' , 'mmole-N/m3' , .false.
To correctly insert this via esm-tools, you can use:
namelist_changes:
namelist_top_cfg:
namtrc:
sn_tracer: "remove_from_namelist"
sn_tracer(1)%clsname: DET
sn_tracer(2)%clsname: ZOO
sn_tracer(3)%clsname: PHY
sn_tracer(1)%cllname: "Detritus"
sn_tracer(2)%cllname: "Zooplankton concentration"
sn_tracer(3)%cllname: "Phytoplankton concentration"
sn_tracer(1:3)%clunit: "mmole-N/m3"
Heterogeneous Parallelization Run (MPI/OpenMP)
Feature available since version: 5.1
In order to run a simulation with hybrid MPI/OpenMP parallelization include the following in your runscript:
Add
heterogenous_parallelization: truein thecomputersection of your runscript. If thecomputersection does not exist create one.Add
omp_num_threads: <number>to the sections of the components you’d like to have OpenMP parallelization.
Example
In AWICM3 we have 3 components: FESOM-2, OpenIFS and RNFMAP. We want to run OpenIFS with 8 OpenMP threads, RNFMAP with 48, and FESOM-2 with 1. Then, the following lines need to be added to our runscript:
general:
[ ... ]
computer:
heterogeneous_parallelization: true
[ ... ]
fesom:
omp_num_threads: 1
[ ... ]
oifs:
omp_num_threads: 8
[ ... ]
rnfmap:
omp_num_threads: 48
[ ... ]
See also
How to setup runscripts for different kind of experiments
This recipe describes how to setup a runscript for the following different kinds of experiments. Besides the variables described in ESM-Tools Variables, add the following variables to your runscript, as described below.
Initial run: An experiment from initial model conditions.
general:
lresume: 0
Restart: An experiment that restarts from a previous experiment with the same experiment id.
general:
lresume: 1
Branching off: An experiment that restarts from a previous experiment but with a different experiment id.
general:
lresume: 1
ini_parent_exp_id: <old-experiment-id>
ini_restart_dir: <path-to-restart-dir-of-old-experiment>/restart/
Branching off and redate: An experiment that restarts from a previous experiment with a different experiment id and if this experiment should be continued with a diiferent start date.
general:
lresume: 1
ini_parent_exp_id: <old-experiment-id>
ini_restart_dir: <path-to-restart-dir-of-old-experiment>/restart/
first_initial_year: <year>
See also
Implement a New Model
Feature available since version: 4.2
Note
since version 6.20.2 a template is available in
esm_tools/configs/templates/component_template.yaml
Upload your model into a repository such us gitlab.awi.de, gitlab.dkrz.de or GitHub. Make sure to set up the right access permissions, so that you comply with the licensing of the software you are uploading.
If you are interested in implementing more than one version of the model, we recommend you to commit them to the master branch in the order they were developed, and that you create a tag per version. For example:
Clone the empty master branch you just created and add your model files to it:
$ git clone https://<your_repository> $ cp -rf <your_model_files_for_given_version> <your_repository_folder> $ git add .
Commit, tag the version and push the changes to your repository:
$ git commit -m "your comment here" $ git tag -a <version_id> -m "your comment about the version" $ git push -u origin <your_master_branch> $ git push origin <version_id>
Repeat steps a and b for all the versions that you would like to be present in ESM-Tools.
Now that you have your model in a repository you are ready to implement it into esm_tools. First, you will need to create your own branch of esm_tools, following the steps 1-4 in Contribution to esm_tools Package. The recommended name for the branch would be
feature/<name_of_your_model>.Then you will need to create a folder for your model inside
esm_tools/configs/componentsand create the model’s yaml file:$ mkdir <PATH>/esm_tools/configs/components/<model> $ touch <PATH>/esm_tools/configs/components/<model>/<model>.yaml
Use your favourite text editor to open and edit your
<model>.yamlin theesm_tools/configs/components/<model>folder:$ <your_text_editor> <PATH>/esm_tools/configs/components/<model>/<model>.yaml
Complete the following information about your model:
# YOUR_MODEL YAML CONFIGURATION FILE # model: your_model_name type: type_of_your_model # atmosphere, ocean, etc. version: "the_default_version_of_your_model"
Include the names of the different versions in the
available_versionssection and the compiling information for the default version:[...] available_versions: - "1.0.0" - "1.0.1" - "1.0.2" git-repository: "https://your_repository.git" branch: your_model_branch_in_your_repo install_bins: "path_to_the_binaries_after_comp" comp_command: "your_shell_commands_for_compiling" # You can use the defaults "${defaults.comp_command}" clean_command: "your_shell_commands_for_cleaning" # You can use the defaults "${defaults.clean_command}" executable: your_model_command setup_dir: "${model_dir}" bin_dir: "${setup_dir}/name_of_the_binary"
In the
install_binskey you need to indicate the path inside your model folder where the binaries are compiled to, so that esm_master can find them once compiled. Theavailable_versionskey is needed for esm_master to list the versions of your model. Thecomp_commandkey indicates the command needed to compile your model, and can be set as${defaults.comp_command}for a default command (mkdir -p build; cd build; cmake ..; make install -j `nproc --all`), or you can define your own list of compiling commands separated with;("command1; command2").At this point you can choose between including all the version information inside the same
<model>.yamlfile, or to distribute this information among different version files:In the
<model>.yaml, use achoose_switch (see Switches (choose_)) to modify the default information that you added in step 7 to meet the requirements for each specific version. For example, each different version has its own git branch:choose_version: "1.0.0": branch: "1.0.0" "1.0.1": branch: "1.0.1" "1.0.2": branch: "develop"
Create a yaml file per version or group of versions. The name of these files should be the same as the ones in the
available_versionssection, in the main<model>.yamlfile or, in the case of a file containing a group of versions, the shared name among the versions (i.e.fesom-2.0.yaml):$ touch <PATH>/esm_tools/configs/<model>/<model-version>.yaml
Open the version file with your favourite editor and include the version specific changes. For example, you want that the version
1.0.2from your model pulls from thedevelopgit branch, instead of from the default branch. Then you add to the<model>-1.0.2.yamlversion file:branch: "develop"
Another example is the
fesom-2.0.yaml. Whilefesom.yamlneeds to contain allavailable_versions, the version specific changes are split amongfesom.yaml(including information about versions 1) andfesom-2.0.yaml(including information about versions 2):[ ... ] available_versions: - '2.0-o' - '2.0-esm-interface' - '1.4' - '1.4-recom' - '1.4-recom-awicm' - '2.0-esm-interface-yac' - '2.0-paleodyn' - '2.0' - '2.0-r' # OG: temporarily here choose_version: '1.4-recom-awicm': branch: fesom_recom_1.4_master destination: fesom-1.4 '1.4-recom': branch: fesom_recom_1.4_master destination: fesom-1.4 [ ... ]
[ ... ] choose_version: '2.0': branch: 2.0.2 git-repository: - https://gitlab.dkrz.de/FESOM/fesom2.git - github.com/FESOM/fesom2.git install_bins: bin/fesom.x 2.0-esm-interface: branch: fesom2_using_esm-interface destination: fesom-2.0 git-repository: - https://gitlab.dkrz.de/a270089/fesom-2.0_yac.git install_bins: bin/fesom.x [ ... ]
Note
These are just examples of model configurations, but the parser used by ESM-Tools to read yaml files (esm_parser) allows for a lot of flexibility in their configuration; i.e., imagine that the different versions of your model are in different repositories, instead of in different branches, and their paths to the binaries are also different. Then you can include the
git-repositoryandinstall_binsvariables inside the corresponding version case for thechoose_version.You can now check if esm_master can list and install your model correctly:
$ esm_masterThis command should return, without errors, a list of available models and versions including yours. Then you can actually try installing your model in the desired folder:
$ mkdir ~/model_codes $ cd ~/model_codes $ esm_master install-your_model-version
If everything works correctly you can check that your changes pass
flake8:$ flake8 <PATH>/esm_tools/configs/components/<model>/<model>.yaml
Use this link to learn more about
flake8and how to install it.Commit your changes, push them to the
originremote repository and submit a pull request through GitHub (see steps 5-7 in Contribution to esm_tools Package).
Note
You can include all the compiling information inside a compile_infos section to avoid
conflicts with other choose_version switches present in your configuration file.
Implement a New Coupled Setup
Feature available since version: 4.2
An example of the different files needed for AWICM setup is included at the end of this section (see Example).
Make sure the models, couplers and versions you want to use, are already available for esm_master to install them (
$ esm_masterand check the list). If something is missing you will need to add it following the instructions in Implement a New Model.Once everything you need is available to esm_master, you will need to create your own branch of esm_tools, following the steps 1-4 in Contribution to esm_tools Package.
Setups need two types of files: 1) coupling files containing information about model versions and coupling changes, and 2) setup files containing the general information about the setup and the model changes. In this step we focus on the creation of the coupling files.
Create a folder for your couplings in
esm_tools/configs/couplings:$ cd esm_tools/configs/couplings/ $ mkdir <coupling_name1> $ mkdir <coupling_name2> ...
The naming convention we follow for the coupling files is
component1-version+component2-version+....Create a yaml file inside the coupling folder with the same name:
$ touch <coupling_name1>/<coupling_name1>.yaml
Include the following information in each coupling file:
components: - "model1-version" - "model2-version" - [ ... ] - "coupler-version" coupling_changes: - sed -i '/MODEL1_PARAMETER/s/OFF/ON/g' model1-1.0/file_to_change - sed -i '/MODEL2_PARAMETER/s/OFF/ON/g' model2-1.0/file_to_change - [ ... ]
The
componentssection should list the models and couplers used for the given coupling including, their required version. Thecoupling_changessubsection should include a list of commands to make the necessary changes in the component’s compilation configuration files (CMakeLists.txt,configure, etc.), for a correct compilation of the coupled setup.
Now, it is the turn for the creation of the setup file. Create a folder for your coupled setup inside
esm_tools/configs/setupsfolder, and create a yaml file for your setup:$ mkdir <PATH>/esm_tools/configs/setups/<your_setup> $ touch <PATH>/esm_tools/configs/setups/<your_setup>/<setup>.yaml
Use your favourite text editor to open and edit your
<setup>.yamlin theesm_tools/configs/setups/<your_setup>folder:$ <your_text_editor> <PATH>/esm_tools/configs/setups/<your_setup>/<setup>.yaml
Complete the following information about your setup:
######################################################################################### ######################### NAME_VERSION YAML CONFIGURATION FILE ########################## ######################################################################################### general: model: your_setup version: "your_setup_version" coupled_setup: True include_models: # List of models, couplers and componentes of the setup. - component_1 # Do not include the version number - component_2 - [ ... ]
Note
Models do not have a
generalsection but in the setups thegeneralsection is mandatory.Include the names of the different versions in the
available_versionssection:general: [ ... ] available_versions: - "1.0.0" - "1.0.1"
The
available_versionskey is needed for esm_master to list the versions of your setup.In the
<setup>.yaml, use achoose_switch (see Switches (choose_)) to assign the coupling files (created in step 3) to their corresponding setup versions:general: [ ... ] choose_version: "1.0.0": couplings: - "model1-1.0+model2-1.0" "1.0.1": couplings: - "model1-1.1+model2-1.1" [ ... ]
You can now check if esm_master can list and install your coupled setup correctly:
$ esm_masterThis command should return, without errors, a list of available setups and versions including yours. Then you can actually try installing your setup in the desire folder:
$ mkdir ~/model_codes $ cd ~/model_codes $ esm_master install-your_setup-version
If everything works correctly you can check that your changes pass
flake8:$ flake8 <PATH>/esm_tools/configs/setups/<your_setup>/<setup>.yaml $ flake8 <PATH>/esm_tools/configs/couplings/<coupling_name>/<coupling_name>.yaml
Use this link to learn more about
flake8and how to install it.Commit your changes, push them to the
originremote repository and submit a pull request through GitHub (see steps 5-7 in Contribution to esm_tools Package).
Example
Here you can have a look at relevant snippets of some of the AWICM-1.0 files.
One of the coupling files for AWICM-1.0 (
esm_tools/configs/couplings/fesom-1.4+echam-6.3.04p1/fesom-1.4+echam-6.3.04p1.yaml):
components:
- echam-6.3.04p1
- fesom-1.4
- oasis3mct-2.8
coupling_changes:
- sed -i '/FESOM_COUPLED/s/OFF/ON/g' fesom-1.4/CMakeLists.txt
- sed -i '/ECHAM6_COUPLED/s/OFF/ON/g' echam-6.3.04p1/CMakeLists.txt
Setup file for AWICM (esm_tools/configs/setups/awicm/awicm.yaml):
#########################################################################################
######################### AWICM 1 YAML CONFIGURATION FILE ###############################
#########################################################################################
general:
model: awicm
#model_dir: ${esm_master_dir}/awicm-${version}
coupled_setup: True
include_models:
- echam
- fesom
- oasis3mct
version: "1.1"
scenario: "PI-CTRL"
resolution: ${echam.resolution}_${fesom.resolution}
postprocessing: false
post_time: "00:05:00"
choose_general.resolution:
T63_CORE2:
compute_time: "02:00:00"
T63_REF87K:
compute_time: "02:00:00"
T63_REF:
compute_time: "02:00:00"
available_versions:
- '1.0'
- '1.0-recom'
- CMIP6
choose_version:
'1.0':
couplings:
- fesom-1.4+echam-6.3.04p1
'1.0-recom':
couplings:
- fesom-1.4+recom-2.0+echam-6.3.04p1
CMIP6:
couplings:
- fesom-1.4+echam-6.3.04p1
Implement a New HPC Machine
To implement a new HPC machine to ESM-Tools, two files need to be updated and created, respectively:
<PATH>/esm_tools/configs/machines/all_machines.yaml<PATH>/esm_tools/configs/machines/<new_machine>.yaml
Add an additional entry for the new machine.
Use your favourite text editor and open the file
<PATH>/esm_tools/configs/machines/all_machines.yaml:$ <your_text_editor> <PATH>/esm_tools/configs/machines/all_machines.yaml
and add a new entry for the new machine (replace placeholders indicated by <…>)
<new_machine>: login_nodes: '<hostname>*' # A regex pattern that matches the hostname of login nodes compute_nodes: '<compute_notes>' # A regex pattern that matches the hostname of compute nodes
Create a new machine file.
Use your favourite text editor to create and edit a new machine file
<new_machine>.yamlin theesm_tools/configs/machines/folder:$ <your_text_editor> <PATH>/esm_tools/configs/machines/<new_machine>.yaml
A template file (
machine_template.yaml) is available inconfigs/templates, so you can alternatively copy this file into theconfigs/machinesfolder edit the relevant entries:$ cp <PATH>/esm_tools/configs/templates/machine_template.yaml <PATH>/esm_tools/configs/machines/<new_machine>.yaml $ <your_text_editor> <PATH>/esm_tools/configs/machines/<new_machine>.yaml
You can also reproduce the two steps above simply by running the following
esm_toolscommand:$ esm_tools create-new-config <PATH>/esm_tools/configs/machines/<new_machine>.yaml -t machine
This will copy the
machine_template.yamlin the target location and open the file in your default editor.
Include a New Forcing/Input File
Feature available since version: 4.2
There are several ways of including a new forcing or input file into your experiment
depending on the degree of control you’d like to achieve. An important clarification is
that <forcing/input>_sources file dictionary specifies the sources (paths to
the files in the pools or personal folders, that need to be copied or linked into the
experiment folder). On the other hand <forcing/input>_files specifies which of these
sources are to be included in the experiment. This allows us to have many sources
already available to the user, and then the user can simply choose which of them to use
by chosing from <forcing/input>_files. <forcing/input>_in_work is used to copy
the files into the work folder (<base_dir>/<exp_id>/run_<DATE>/work) if necessary
and change their name. For more technical details see File Dictionaries.
The next sections illustrate some of the many options to handle forcing and input files.
Source Path Already Defined in a Config File
Make sure the source of the file is already specified inside the
forcing_sourcesorinput_sourcesfile dictionaries in the configuration file of the setup or model you are running, or on thefurther_readingfiles.In your runscript, include the key of the source file you want to include inside the
forcing_filesorinput_filessection.
Note
Note that the key containing the source in the forcing_sources or
input_sources can be different than the key specified in forcing_files or
input_files.
Example
In ECHAM, the source and input file paths are specified in a separate file
(<PATH>/esm_tools/configs/components/echam/echam.datasets.yaml) that
is reached through the further_reading section of the echam.yaml. This
file includes a large number of different sources for input and forcing contained
in the pool directories of the HPC systems Ollie and Mistral. Let’s have a look
at the sst forcing file options available in this file:
forcing_sources:
# sst
"amipsst":
"${forcing_dir}/amip/${resolution}_amipsst_@YEAR@.nc":
from: 1870
to: 2016
"pisst": "${forcing_dir}/${resolution}${ocean_resolution}_piControl-LR_sst_1880-2379.ncy"
This means that from our runscript we will be able to select either amipsst
or pisst as sst forcing files. If you define scenario in ECHAM be
PI-CTRL the correct file source (pisst) is already selected for you.
However, if you would like to select this file manually you can just simply add
the following to your runscript:
forcing_files:
sst: pisst
Modify the Source of a File
To change the path of the source for a given forcing or input file from your runscript:
Include the source path under a key inside
forcing_sourcesorinput_sourcesin your runscript:<forcing/input>_sources: <key_for_your_file>: <path_to_your_file>
If the source is not a single file, but there is a file per year use the
@YEAR@andfrom:to:functionality in the path to copy only the files corresponding to that run’s year:<forcing/input>_sources: <key_for_your_source>: <firt_part_of_the_path>@YEAR@<second_part_of_the_path> from: <first_year> to: <last_year>
Make sure the key for your path is defined in one of the config files that you are using, inside of either
forcing_filesorinput_files. If it is not defined anywhere you will have to include it in your runscript:<forcing/input>_files: <key_for_your_file>: <key_for_your_source>
Copy the file in the work folder and/or rename it
To copy the files from the forcing/input folders into the work folder
(<base_dir>/<exp_id>/run_<DATE>/work) or rename them:
Make sure your file and its source is defined somewhere (either in the config files or in your runscript) in
<forcing/input>_sourcesand<forcing/input>_files(see subsections Source Path Already Defined in a Config File and Modify the Source of a File).In your runscript, add the key to the file you want to copy with value the same as the key, inside <forcing/input>_in_work:
<forcing/input>_in_work: <key_for_your_file>: <key_for_your_file>
If you want to rename the file set the value to the desired name:
<forcing/input>_in_work: <key_for_your_file>: <key_for_your_file>
Example
In ECHAM the sst forcing file depends in the scenario defined by the user:
esm_tools/config/component/echam/echam.datasets.yaml
forcing_sources:
# sst
"amipsst":
"${forcing_dir}/amip/${resolution}_amipsst_@YEAR@.nc":
from: 1870
to: 2016
"pisst": "${forcing_dir}/${resolution}${ocean_resolution}_piControl-LR_sst_1880-2379.nc"
esm_tools/config/component/echam/echam.yaml
choose_scenario:
"PI-CTRL":
forcing_files:
sst: pisst
[ ... ]
If scenario: "PI-CTRL" then the source selected will be
${forcing_dir}/${resolution}${ocean_resolution}_piControl-LR_sst_1880-2379.nc
and the name of the file copied to the experiment forcing folder will be
${resolution}${ocean_resolution}_piControl-LR_sst_1880-2379.nc. However,
ECHAM needs this file in the same folder as the binary (the work folder)
under the name unit.20. To copy and rename this file into the work folder
the following lines are used in the echam.yaml configuration file:
forcing_in_work:
sst: "unit.20"
You can use the same syntax inside your runscript to copy into the work
folder any forcing or input file, and rename it.
See also
Exclude a Forcing/Input File
Feature available since version: 4.2
To exclude one of the predefined forcing or input files from being copied to your experiment folder:
Find the key of the file to be excluded inside the config file,
<forcing/input>_filesfile dictionary.In your runscript, use the
remove_functionality to exclude this key from the<forcing/input>_filesfile dictionary:remove_<input/forcing>_files: - <key_of_the_file1> - <key_of_the_file2> - ...
Example
To exclude the sst forcing file from been copied to the experiment folder
include the following lines in your runscript:
remove_forcing_files:
- sst
Using your own namelist
Feature available since version: 4.2
Warning
This feature is only recommended if the number of changes that need to be applied to the default
namelist is very large, otherwise we recommend to use the feature namelist_changes (see
Changing Namelist Entries from the Runscript). You can check the default namelists here.
In your runscript, you can instruct ESM-Tools to substitute a given default namelist by a namelist of your choice.
Search for the
config_sourcesvariable inside the configuration file of the model you are trying to run, and then, identify the “key” containing the path to the default namelist.In your runscript, indented in the corresponding model section, add an
add_config_sourcessection, containing a variable whose “key” is the one of step 1, and the value is the path of the new namelist.Bare in mind, that namelists are first loaded by ESM-Tools, and then modified by the default
namelist_changesin the configuration files. If you want to ignore all those changes for the your new namelist you’ll need to addremove_namelist_changes: [<name_of_your_namelist>].
In dot notation both steps will look like:
<model_name>.<add_config_sources>.<key_of_the_namelist>: <path_of_your_namelist>
<model_name>.<remove_namelis_changes>: [<name_of_your_namelist>]
Warning
Use step 3 at your own risk! Many of the model specific information and functionality is
transferred to the model through namelist_changes, and therefore, we discourage you from using
remove_namelist_changes unless you have a very deep understanding of the configuration file and the model.
Following Changing Namelist Entries from the Runscript would be a safest solution.
Example
In this example we show how to use an ECHAM namelist.echam and a FESOM namelist.ice that are not
the default ones and omit the namelist_changes present in echam.yaml and fesom.yaml configuration
files.
Following step 1, search for the config_sources dictionary inside the echam.yaml:
# Configuration Files:
config_sources:
"namelist.echam": "${namelist_dir}/namelist.echam"
In this case the “key” is "namelist.echam" and the “value” is "${namelist_dir}/namelist.echam".
Let’s assume your namelist is in the directory /home/ollie/<usr>/my_namelists. Following step 2,
you will need to include the following in your runscript:
echam:
add_config_sources:
"namelist.echam": /home/ollie/<usr>/my_namelists/namelist.echam
If you want to omit the namelist_changes in echam.yaml or any other configuration file
that your model/couple setup is using, you’ll need to add to your runscript
remove_namelist_changes: [namelist.echam] (step 3):
echam:
add_config_sources:
"namelist.echam": /home/ollie/<usr>/my_namelists/namelist.echam
remove_namelist_changes: [namelist.echam]
Warning
Many of the model specific information and functionality is transferred to the model
through namelist_changes, and therefore, we discourage you from using this unless you
have a very deep understanding of the echam.yaml file and the ECHAM model. For example,
using remove_namelist_changes: [namelist.echam] will destroy the following lines in the
echam.yaml:
choose_lresume:
False:
restart_in_modifications:
"[[streams-->STREAM]]":
- "vdate <--set_global_attr-- ${start_date!syear!smonth!sday}"
# - fdate "<--set_dim--" ${year_before_date}
# - ndate "<--set_dim--" ${steps_in_year_before}
True:
# pseudo_start_date: $(( ${start_date} - ${time_step} ))
add_namelist_changes:
namelist.echam:
runctl:
dt_start: "remove_from_namelist"
This lines are relevant for correctly performing restarts, so if
remove_namelist_changes is used, make sure to have the approrpiate commands on your
runscript to remove dt_start from your namelist in case of a restart.
Following step 1, search for the config_sources dictionary inside the fesom.yaml:
config_sources:
config: "${namelist_dir}/namelist.config"
forcing: "${namelist_dir}/namelist.forcing"
ice: "${namelist_dir}/namelist.ice"
oce: "${namelist_dir}/namelist.oce"
diag: "${namelist_dir}/namelist.diag"
In this case the “key” is ice and the “value” is ${namelist_dir}/namelist.ice.
Let’s assume your namelist is in the directory /home/ollie/<usr>/my_namelists. Following step 2,
you will need to include the following in your runscript:
fesom:
add_config_sources:
ice: "/home/ollie/<usr>/my_namelists/namelist.ice"
If you want to omit the namelist_changes in fesom.yaml or any other configuration file
that your model/couple setup is using, you’ll need to add to your runscript
remove_namelist_changes: [namelist.ice] (step 3):
fesom:
add_config_sources:
ice: "/home/ollie/<usr>/my_namelists/namelist.ice"
remove_namelist_changes: [namelist.ice]
Warning
Many of the model specific information and functionality is transferred to the model
through namelist_changes, and therefore, we discourage you from using this unless you
have a very deep understanding of the fesom.yaml file and the FESOM model.
How to branch-off FESOM from old spinup restart files
When you branch-off from very old FESOM ocean restart files, you may encounter the following runtime error:
read ocean restart file
Error:
NetCDF: Invalid dimension ID or name
This is because the naming of the NetCDF time dimension variable in the restart file has changed from T to time during the development of FESOM and the different FESOM versions.
Therefore, recent versions of FESOM expect the name of the time dimension to be time.
In order to branch-off experiments from spinup restart files that use the old name for the time dimension, you need to rename this dimension before starting the branch-off experiment.
Warning
The following work around will change the restart file permanently. Make sure you do not apply this to the original file.
To rename a dimension variable of a NetCDF file, you can use ncrename:
ncrename -d T,time <copy_of_restart_spinup_file>.nc
where T is the old dimension and time is the new dimension.
See also
cookbook:How to run a branch-off experiment
Recieve batch notifications via e-mail
It is possible to define the following variables in your runscript either in computer or in general (up to you which one) to recieve e-mail notifications about the status of your job:
general/computer:
mail_type: <type_of_notification>
mail_user: <your_e-mail>
Find more about the possible values of mail_type in SLURM documentation (https://slurm.schedmd.com/sbatch.html#OPT_mail-type)